Publications
Lab Notes
| BIOCHEMISTRY Uncoiling DNA Analysis |
Posted November 2010 |
A rapid DNA-fingerprinting method would identify biological organisms in the field. |
|
DNA sequencing has proven to be a useful tool for identifying organisms, but is not currently able to perform rapid identification of potential bioagents from mixed samples in the field. Now Lincoln Laboratory researchers Lalitha Parameswaran, an electrical engineer, and Eric Schwoebel, a biologist, are working to build a portable, rapid DNA fingerprinting device that could examine samples quickly and recognize a wide variety of organisms.
The core of the technique is restriction enzymes, which are produced naturally in bacteria and protect their hosts by binding to specific known sequences on foreign DNA strands and cutting the strands into fragments. Schwoebel, a member of the Laboratory's Chemical, Biological, and Nanoscale Technologies Group, recognized that if they could get the restriction enzymes to bind but not cut the DNA and attach fluorescent markers to the enzymes, they should be able to create optically detectable patterns of binding, or "fingerprints" specific to a particular organism. Essentially they would get a string of glowing markers, arranged one way for, say, anthrax and another for E. coli.
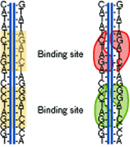 Restriction enzymes attach at specific locations on DNA. They can be illuminated with optical tags.
Restriction enzymes attach at specific locations on DNA. They can be illuminated with optical tags."We want the restriction enzymes to bind to the DNA but retain their sequence specificity," says Parameswaran. "We already have sequence information for many organisms of interest, so we know the binding pattern that will be produced by each restriction enzyme. With this technique, we may not need to completely sequence unknown target DNA in order to identify its host organism."
There are, of course, existing approaches to DNA identification. Researchers can use the polymerase chain reaction (PCR) to create multiple copies of a strand of DNA and real-time PCR to produce a fluorescent signal indicating the amount of target-specific DNA produced. The DNA can also be sequenced by using one of several standard methods. But these approaches have many time- and labor-intensive steps, and can also require a very clean sample, necessitating stringent sample-preparation procedures to remove inhibitory components.
The team's proposal, in contrast, would use a much simpler assay. It could use a less pure sample, say from the soil or a drop of blood, or even a suspicious film scraped off a building's window if someone suspects that a terrorist has fogged the building with an airborne pathogen. The advantage of the method is that it doesn't require isolating the specific target DNA to be identified, or even foreknowledge of what target is present in the sample. One could simply extract and fingerprint all of the DNA in a sample, and the fingerprints could then be matched using already existing sequence databases.
The trick is to coax the restriction enzymes to bind to DNA sequences without actually cutting them. The pair says they have done that by altering the salts in a solution containing the DNA. The enzymes rely on magnesium ions to enable the cutting reaction, but replacing the magnesium with calcium stops them from snipping the strand. Eventually, they'd like to use restriction enzymes that have the cutting action permanently suppressed. These would be produced from genetically altered bacteria, something researchers elsewhere are working on. One issue the team is studying is whether they can interfere with the cutting and yet keep the enzymes binding to the right sites.
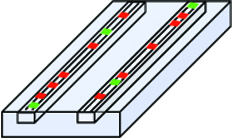
Another key requirement of the detection system is to create tiny channels, only a few nanometers wide, on a sample plate. An applied electric field would draw the DNA strands with bound restriction enzymes through the channels while an optical system watched for the signal from the fluorescent tags. The channels keep the DNA strand stretched out and localized; if it were to fold back on itself or ball up, it would be impossible to see the linear pattern of binding sites that identified a particular organism. Creating channels only 10 nanometers wide, or even narrower, presents an engineering challenge, but not an insurmountable one, the team believes.
 |
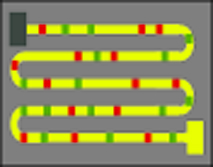 |
One critical part of this procedure is the straightening out of the spaghetti-like labeled DNA strand. Once it is laid out in nanochannels, shown on the left, the entire strand can be imaged, as shown on the right, to give a binding pattern signature unique to the organism from which the DNA originated.. |
|
The plan will also require a sophisticated optical system to detect small amounts of fluorescence in microseconds, as well as a computer to compare any patterns detected to known DNA sequences. Eventually, they'd like a system that uses a sample plate with hundreds or thousands of channels for high throughput. "It would be analogous to analyzing the amount of DNA present in the entire human genome in a matter of minutes," she says.
One plus of the scheme is that it could provide information about an unknown or genetically modified organism. "Even if you haven't seen this new Bacillus organism before, you might still be able to tell it's a Bacillus," Schwoebel says. It might be possible to find patterns that showed a particular strain of bacteria was resistant to a certain antibiotic so that doctors could prescribe a different one to people who had been exposed, "It could give you an idea of how to proceed without having to go through cell-culture techniques to identify a new organism," he says.
In addition to providing quick, in-the-field information in a possible biological attack, the rapid identification scheme could also prove useful in medicine for bedside diagnosis and personalized medicine. It might also be helpful in food safety and forensic science.
The team's current focus is on testing their ideas about non-destructive specific binding of the enzymes, a project they hope to complete by the end of the year. If they're successful, they hope to find funding to continue the development of a full system.